NicheNet 이해하기
추천글 : 【생물정보학】 리간드-수용체 상호작용 분석
1. 개요 [본문]
2. 데이터셋 [본문]
3. 알고리즘 [본문]
4. 결론 [본문]
1. 개요 [목차]
⑴ 경로 활성화를 식별하고 관련된 잠재적인 리간드를 강조하는 툴
⑵ 즉, 특정 DEG가 있을 때 어떤 cell 혹은 어떤 ligand가 관련 있는지 구하는 것
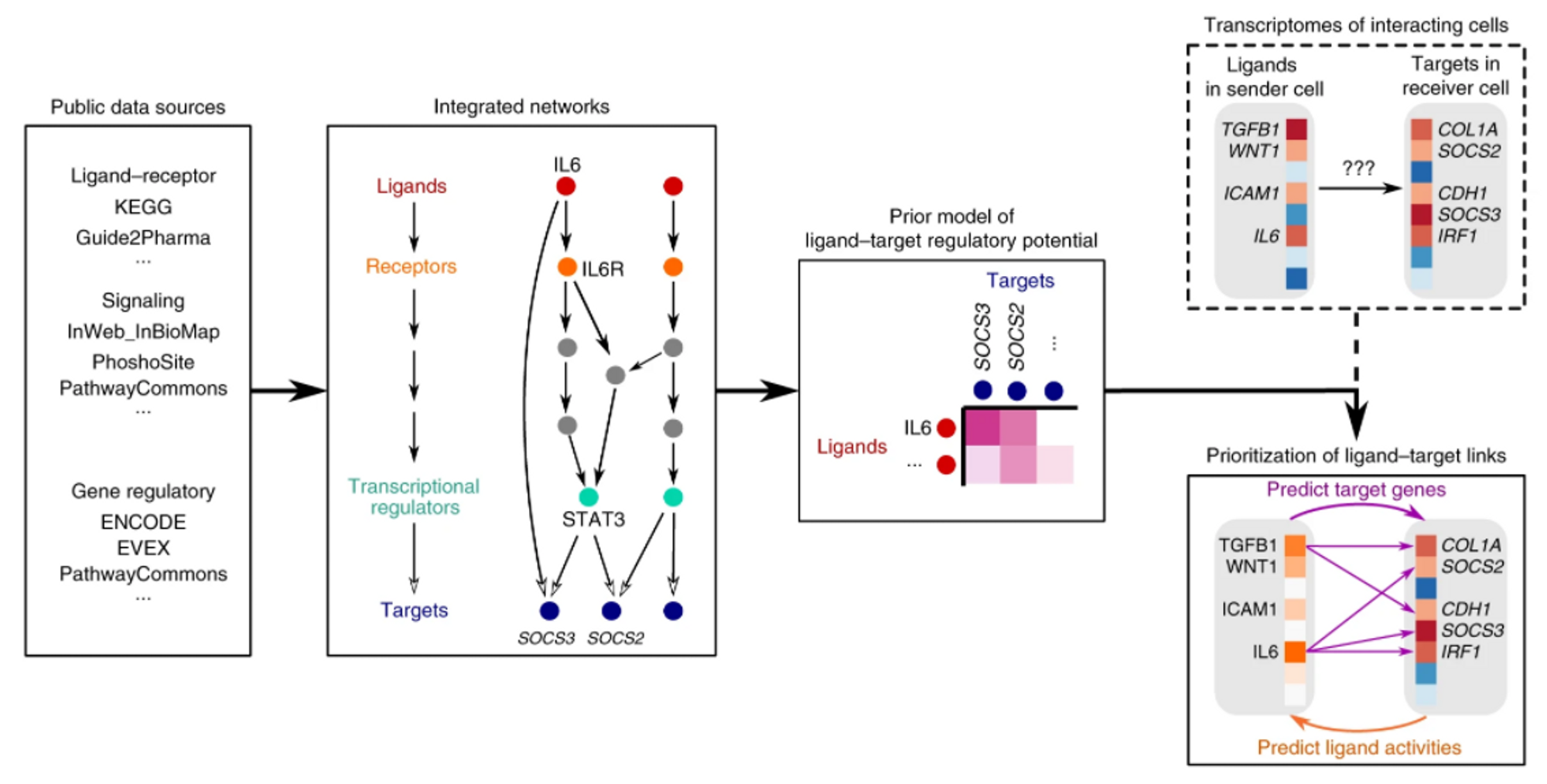
⑶ 모식도 : 여러 데이터셋을 그래프 구조로 이어서 분석
Figure. 1. NicheNet 모식도
⑷ 코드 : https://github.com/saeyslab/nichenetr
2. 데이터셋 [목차]
⑴ 종합
① cpdb
○ cpdb_interaction : Kamburov et al. (2013)
○ cpdb_complex : Kamburov et al. (2013)
② evex
○ lr_evex_regulation_expression : Van Landeghem et al. (2012)
○ evex_regulation_expression : Van Landeghem et al. (2012)
○ evex_catalysis : Van Landeghem et al. (2012)
○ evex_regulation_other : Van Landeghem et al. (2012)
○ evex_phosphorylation : Van Landeghem et al. (2012)
○ evex_regulation_binding : Van Landeghem et al. (2012)
○ evex_binding : Van Landeghem et al. (2012)
③ guide2pharmacology
○ pharmacology : Pawson et al. (2014)
④ harmonizome
○ harmonizome_KEA : Lachmann and Ma’ayan (2009)
○ harmonizome_PhosphoSite : Hornbeck et al. (2015)
○ harmonizome_kinase_substrate_predictions : Rouillard et al. (2016)
○ harmonizome_DEPOD : Duan et al. (2015)
○ harmonizome_CHEA : Lachmann et al. (2010)
○ harmonizome_ENCODE : Consortium (2004)
○ harmonizome_JASPAR : Mathelier et al. (2014)
○ harmonizome_TRANSFAC_CUR : Matys et al. (2006)
○ harmonizome_TRANSFAC : Matys et al. (2006)
○ harmonizome_MOTIFMAP : Xie et al. (2009)
○ harmonizome_GEO_TF : Edgar et al. (2002)
○ harmonizome_GEO_KINASE : Edgar et al. (2002)
○ harmonizome_GEO_GENE : Edgar et al. (2002)
○ harmonizome_MSIGDB_GENE : Subramanian et al. (2005)
⑤ HTRIDB
○ HTRIDB : Bovolenta et al. (2012)
⑥ inweb_inbiomap
○ inweb_inbio_interaction : Li et al. (2017)
○ inweb_inbio_interaction_pathway : Li et al. (2017)
○ inweb_inbio_pathway : Li et al. (2017)
⑦ kegg
○ kegg_cytokines : Kanehisa et al. (2016)
○ kegg_cams : Kanehisa et al. (2016)
○ kegg_neuroactive : Kanehisa et al. (2016)
○ kegg_ecm : Kanehisa et al. (2016)
⑧ omnipath
○ omnipath_directional : Türei et al. (2016)
○ omnipath_undirectional : Türei et al. (2016)
⑨ ontogenet
○ ontogenet_coarse : Jojic et al. (2013)
⑩ pathwaycommons
○ lr_pathwaycommons_controls_expression_of : Cerami et al. (2011)
○ pathwaycommons_controls_expression_of : Cerami et al. (2011)
○ pathwaycommons_controls_phosphorylation_of : Cerami et al. (2011)
○ pathwaycommons_controls_state_change_of : Cerami et al. (2011)
○ pathwaycommons_catalysis_precedes : Cerami et al. (2011)
○ pathwaycommons_controls_transport_of : Cerami et al. (2011)
○ pathwaycommons_interacts_with : Cerami et al. (2011)
○ pathwaycommons_in_complex_with : Cerami et al. (2011)
⑪ ppi
○ ppi_lr
○ ppi_l_bidir
○ ppi_bidir_r
○ ppi_bidir_bidir
○ ppi_lr_go
○ ppi_l_bidir_go
○ ppi_bidir_r_go
○ ppi_bidir_bidir_go
⑫ ramilowski
○ ramilowski_known : Ramilowski et al. (2015)
⑬ regnetwork
○ regnetwork_source : Liu et al. (2015)
○ regnetwork_encode : Liu et al. (2015)
⑭ Remap
○ Remap_5 : Griffon et al. (2015)
⑮ trrust
○ trrust : Han et al. (2015)
⑯ vinayagam
○ vinayagam_ppi : Vinayagam et al. (2011)
⑵ 종류 2. protein-protein interaction
① Omnipath
② PathwayCommons
③ InWeb
④ ConsensusPathDB
⑤ Vinayagam et al
⑥ EVEX
⑦ KEA
⑧ PhosphoSite
⑨ DEPOD
⑩ Harmonizome kinase–substrate predictions
⑶ 종류 3. gene regulatory interaction
① RegNetwork
② TRRUST
③ HTRIDB
④ ReMap
⑤ EVEX
⑥ PathwayCommons
⑦ Ontogenet
⑧ CHEA
⑨ ENCODE
⑩ JASPAR
⑪ TRANSFAC
⑫ MOTIFMAP
⑬ Gene Expression Omnibus (GEO)
⑭ MSigDBHarmonizome
3. 알고리즘 [목차]
⑴ 단계 1. 한 개의 통합 네트워크가 아니라 두 개의 네트워크로 진행
① 종류 1. protein–protein interaction for ligand–receptor and signaling data sources
② 종류 2. gene regulatory interaction in the gene regulatory network
⑵ 단계 2. weighted sum of adjacency matrices

⑶ 단계 3. mlrMBO를 이용한 parameter optimization
① mlrMBO : modular framework for model-based optimization of expensive black-box functions
⑷ 단계 4. weighted matrix를 랜덤하게 생성
⑸ 단계 5. ligand-target regulatory potential score 계산
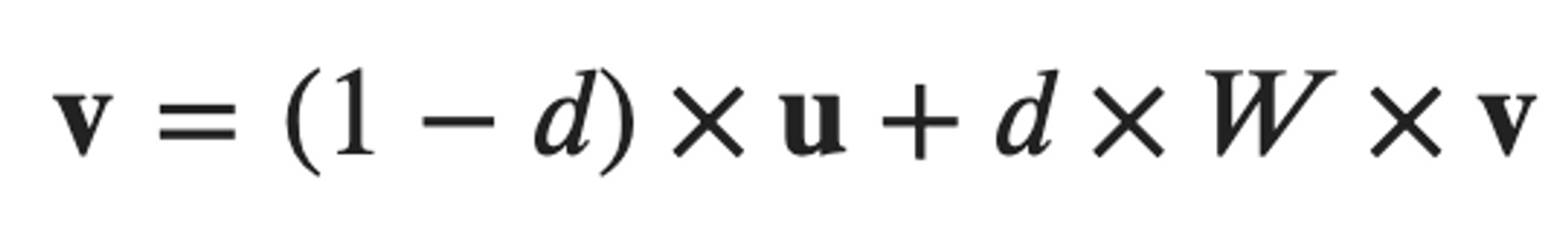
⑹ 단계 6. 모델 생성
① 목적 : 다중 data source 중 하나를 선별하기 위함
② 종류 1. leave-one-in
③ 종류 2. one-versus-one-versus-one
⑺ 단계 7. validation
① target gene prediction 평가
② ligand activity prediction 평가
③ cell type bias 평가
④ IPA(upstream regulator analysis of ingenuity pathway analysis)와의 비교
⑤ CCCExplorer와의 비교
⑻ 단계 8. 다른 데이터셋에서의 적용 및 비주얼라이제이션
4. 결론 [목차]
⑴ CellPhonedB보다 더 좋다는 의견이 있음
입력: 2023.09.15 11:15
'▶ 자연과학 > ▷ 생물정보학' 카테고리의 다른 글
| 【생물정보학】 리간드-수용체 상호작용 분석 (0) | 2016.06.27 |
|---|---|
| 【생물정보학】 세포주 (셀라인) 라이브러리 (0) | 2016.06.27 |
| 【생물정보학】 생물도감 (0) | 2016.06.24 |
| 【생물정보학】 Seurat와 scanpy의 비교 (0) | 2016.06.22 |
| 【생물정보학】 질환 (병리) 라이브러리 (0) | 2016.06.21 |

최근댓글